Differential and Interactive Effects of Substrate Topography and Chemistry on Human Mesenchymal Stem Cell Gene Expression
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characteristics of Material
2.2. Cell Growth and Morphology
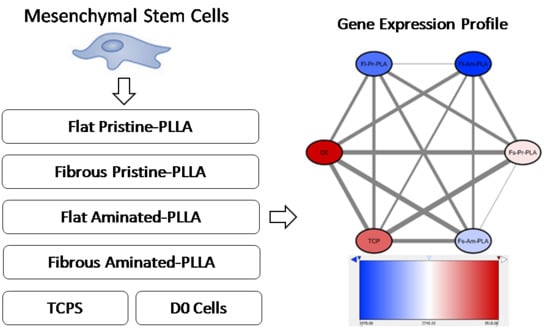
2.3. RNA-Seq Data Analysis
3. Materials and Methods
3.1. Materials
3.2. Study Groups
3.3. Preparation and Characterization of PLLA Substrates
3.4. Cell Culture
3.5. RNA Extraction
3.6. RNA-Seq and Data Analysis
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Availability of data and materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ryu, K.-H.; Cho, K.-A.; Park, H.S.; Kim, J.-Y.; Woo, S.-Y.; Jo, I.; Choi, Y.H.; Park, Y.M.; Jung, S.-C.; Chung, S.M. Tonsil-derived mesenchymal stromal cells: Evaluation of biologic, immunologic and genetic factors for successful banking. Cytotherapy 2012, 14, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Riekstina, U.; Cakstina, I.; Parfejevs, V.; Hoogduijn, M.; Jankovskis, G.; Muiznieks, I.; Muceniece, R.; Ancans, J. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009, 5, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Song, Q.; Shen, J.; Xu, L.; Xu, Z.; Wu, R.; Ge, Y.; Zhu, J.; Wu, J.; Dou, Q. Comparison of human adipose stromal vascular fraction and adipose-derived mesenchymal stem cells for the attenuation of acute renal ischemia/reperfusion injury. Sci. Rep. 2017, 7, 44058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Li, D.; Song, K.; Wei, J.; Yao, S.; Li, Z.; Su, X.; Ju, X.; Chao, L.; Deng, X. Exosomes derived from human umbilical cord mesenchymal stem cells protect against cisplatin-induced ovarian granulosa cell stress and apoptosis in vitro. Sci. Rep. 2017, 7, 2552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjornson, C.R.; Rietze, R.L.; Reynolds, B.A.; Magli, M.C.; Vescovi, A.L. Turning brain into blood: A hematopoietic fate adopted by adult neural stem cells in vivo. Science 1999, 283, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Mezey, E.; Chandross, K.J.; Harta, G.; Maki, R.A.; McKercher, S.R. Turning blood into brain: Cells bearing neuronal antigens generated in vivo from bone marrow. Science 2000, 290, 1779–1782. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.G.; Shuttleworth, C.A.; Kielty, C.M. Mesenchymal stem cells and neovascularization: Role of platelet-derived growth factor receptors. J. Cell. Mol. Med. 2007, 11, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.L.; Zhao, Q.; Zhang, Y.C.; Cheng, L.; Liu, M.; Shi, J.; Yang, Y.Z.; Pan, C.; Ge, J.; Phillips, M.I. Autologous mesenchymal stem cell transplantation induce VEGF and neovascularization in ischemic myocardium. Regul. Pept. 2004, 117, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Lou, R.; Huang, F.; Peng, Y.; Jiang, Z.; Huang, K.; Wu, X.; Zhang, Y.; Fan, Z.; Zhou, H. Immunomodulation effects of mesenchymal stromal cells on acute graft-versus-host disease after hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2015, 21, 97–104. [Google Scholar] [CrossRef] [PubMed]
- English, K. Mechanisms of mesenchymal stromal cell immunomodulation. Immunol. Cell Biol. 2013, 91, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.M.; Rahman, A.A.; Miller, S.; Stavely, R.; Sakkal, S.; Nurgali, K. The neuroprotective effects of human bone marrow mesenchymal stem cells are dose-dependent in TNBS colitis. Stem Cell Res. Ther. 2017, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ren, X.; Li, G.; Jiao, C.; Zhang, L.; Zhao, S.; Wang, J.; Han, Z.C.; Li, X. Mesenchymal stem cells ameliorate experimental autoimmune uveoretinitis by comprehensive modulation of systemic autoimmunity. Invest. Ophthalmol. Vis. Sci. 2011, 52, 3143–3152. [Google Scholar] [CrossRef] [PubMed]
- Mahara, A.; Yamaoka, T. Continuous separation of cells of high osteoblastic differentiation potential from mesenchymal stem cells on an antibody-immobilized column. Biomaterials 2010, 31, 4231–4237. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Abdeen, A.A.; Kilian, K.A. Rewiring mesenchymal stem cell lineage specification by switching the biophysical microenvironment. Sci. Rep. 2014, 4, 5188. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.P.; Hui, T.; Yeung, C.; Li, J.; Mo, I.; Chan, G. Self-assembled collagen–human mesenchymal stem cell microspheres for regenerative medicine. Biomaterials 2007, 28, 4652–4666. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Butler, J.P.; Ingber, D.E. Mechanotransduction across the cell surface and through the cytoskeleton. Science 1993, 260, 1124–1127. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Gao, P.-L.; Wang, K.; Wang, J.-Y.; Zhong, Y.; Luo, Y. Fibrous scaffolds potentiate the paracrine function of mesenchymal stem cells: A new dimension in cell-material interaction. Biomaterials 2017, 141, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Cukierman, E.; Pankov, R.; Stevens, D.R.; Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 2001, 294, 1708–1712. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.-C.; Fujita, S.; Tonami, H.; Kato, K.; Iwata, H.; Hsu, S.-H. Cell orientation and regulation of cell–cell communication in human mesenchymal stem cells on different patterns of electrospun fibers. Biomed. Mater. 2013, 8, 055002. [Google Scholar] [CrossRef] [PubMed]
- Nedjari, S.; Awaja, F.; Altankov, G. Three Dimensional Honeycomb Patterned Fibrinogen Based Nanofibers Induce Substantial Osteogenic Response of Mesenchymal Stem Cells. Sci. Rep. 2017, 7, 15947. [Google Scholar] [CrossRef] [PubMed]
- Amjadian, S.; Seyedjafari, E.; Zeynali, B.; Shabani, I. The synergistic effect of nano-hydroxyapatite and dexamethasone in the fibrous delivery system of gelatin and poly(L-lactide) on the osteogenesis of mesenchymal stem cells. Int. J. Pharm. 2016, 507, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Woo, K.M.; Jun, J.-H.; Chen, V.J.; Seo, J.; Baek, J.-H.; Ryoo, H.-M.; Kim, G.-S.; Somerman, M.J.; Ma, P.X. Nano-fibrous scaffolding promotes osteoblast differentiation and biomineralization. Biomaterials 2007, 28, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ma, T. Perfusion regulation of hMSC microenvironment and osteogenic differentiation in 3D scaffold. Biotechnol. Bioeng. 2012, 109, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Q.; Wen, F.; Chen, H.Z.; Pal, M.; Lai, Y.K.; Zhao, A.Z.; Tan, L.P. Micropatterning Extracellular Matrix Proteins on Electrospun Fibrous Substrate Promote Human Mesenchymal Stem Cell Differentiation Toward Neurogenic Lineage. ACS Appl. Mater. Interfaces 2016, 8, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Bagher, Z.; Ebrahimi-Barough, S.; Azami, M.; Safa, M.; Joghataei, M.T. Cellular activity of Wharton’s Jelly-derived mesenchymal stem cells on electrospun fibrous and solvent-cast film scaffolds. J. Biomed. Mater. Res. A 2016, 104, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Sreerekha, P.R.; Menon, D.; Nair, S.V.; Chennazhi, K.P. Fabrication of electrospun poly (lactide-co-glycolide)–fibrin multiscale scaffold for myocardial regeneration in vitro. Tissue Eng. Part A 2012, 19, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.K.F.; Wan, A.C.A.; Le Visage, C.; Liao, I.C.; Leong, K.W. Proliferation and differentiation of human mesenchymal stem cell encapsulated in polyelectrolyte complexation fibrous scaffold. Biomaterials 2006, 27, 6111–6122. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.E.; Petrie, T.A.; Creighton, F.P.; García, A.J. Human mesenchymal stem cell differentiation on self-assembled monolayers presenting different surface chemistries. Acta Biomater. 2010, 6, 12–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, N.J.; Gentleman, E. Evolving insights in cell–matrix interactions: Elucidating how non-soluble properties of the extracellular niche direct stem cell fate. Acta Biomater. 2015, 11, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.L.; McDevitt, T.C.; Engler, A J. Materials as stem cell regulators. Nat. Mater. 2014, 13, 547. [Google Scholar] [CrossRef] [PubMed]
- Cochis, A.; Grad, S.; Stoddart, M.; Farè, S.; Altomare, L.; Azzimonti, B.; Alini, M.; Rimondini, L. Bioreactor mechanically guided 3D mesenchymal stem cell chondrogenesis using a biocompatible novel thermo-reversible methylcellulose-based hydrogel. Sci. Rep. 2017, 7, 45018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hettiaratchi, M.; Guldberg, R.; McDevitt, T. Biomaterial strategies for controlling stem cell fate via morphogen sequestration. J. Mater. Chem. B 2016, 4, 3464–3481. [Google Scholar] [CrossRef]
- Spees, J.L.; Lee, R.H.; Gregory, C.A. Mechanisms of mesenchymal stem/stromal cell function. Stem Cell Res. Ther. 2016, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, B.; Mekala, N.K.; Baadhe, R.R.; Rao, P.S. Role of signaling pathways in mesenchymal stem cell differentiation. Curr. Stem Cell Res. Ther. 2014, 9, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Tanavde, V.M.; Liew, L.; Lim, J.; Ng, F. Regulatory Networks in Stem Cells; Signaling Networks in Mesenchymal Stem Cells; Humana Press: Totowa, NJ, USA, 2009; pp. 329–335. [Google Scholar]
- Kasoju, N.; Wang, H.; Zhang, B.; George, J.; Gao, S.; Triffitt, J.T.; Cui, Z.; Ye, H. Transcriptomics of human multipotent mesenchymal stromal cells: Retrospective analysis and future prospects. Biotechnol. Adv. 2017, 35, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.H.; Zhou, Y.X.; Lee, M.S.; Zhang, Y.Z.; Li, W.J. A newly identified mechanism involved in regulation of human mesenchymal stem cells by fibrous substrate stiffness. Acta Biomater. 2016, 42, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, A.C.; Liu, Y.J.; Ma, T. Expansion of human mesenchymal stem cells in fibrous bed bioreactor. Biochem. Eng. J. 2016, 108, 51–57. [Google Scholar] [CrossRef]
- Tenney, R.M.; Discher, D.E. Stem cells, microenvironment mechanics, and growth factor activation. Curr. Opin. Cell Biol. 2009, 21, 630–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Mao, Z.; Shi, H.; Gao, C. In-depth study on aminolysis of poly (ɛ-caprolactone): Back to the fundamentals. Sci. China Chem. 2012, 55, 2419–2427. [Google Scholar] [CrossRef]
- Yuan, S.; Xiong, G.; Wang, X.; Zhang, S.; Choong, C. Surface modification of polycaprolactone substrates using collagen-conjugated poly (methacrylic acid) brushes for the regulation of cell proliferation and endothelialisation. J. Mater. Chem. 2012, 22, 13039–13049. [Google Scholar] [CrossRef]
- Zhu, Y.; Gao, C.; Liu, X.; Shen, J. Surface modification of polycaprolactone membrane via aminolysis and biomacromolecule immobilization for promoting cytocompatibility of human endothelial cells. Biomacromolecules 2002, 3, 1312–1319. [Google Scholar] [CrossRef] [PubMed]
- Asti, A.; Gioglio, L. Natural and synthetic biodegradable polymers: Different scaffolds for cell expansion and tissue formation. Int. J. Artif. Organs 2014, 37, 187–205. [Google Scholar] [PubMed]
- Anderson, H.J.; Sahoo, J.K.; Ulijn, R.V.; Dalby, M.J. Mesenchymal stem cell fate: Applying biomaterials for control of stem cell behavior. Front. Bioeng. Biotechnol. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Nalluri, S.M.; Hill, M.J.; Sarkar, D. Essentials of Mesenchymal Stem Cell Biology and Its Clinical Translation; Control of Mesenchymal Stem Cells with Biomaterials; Springer: Dordrecht, The Netherlands, 2013; pp. 139–159. [Google Scholar]
- Benoit, D.S.; Schwartz, M.P.; Durney, A.R.; Anseth, K.S. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat. Mater. 2008, 7, 816–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakian, R.; Block, T.J.; Johnson, S.M.; Marinkovic, M.; Wu, J.; Dai, Q.; Dean, D.D.; Chen, X.-D. Native extracellular matrix preserves mesenchymal stem cell “stemness” and differentiation potential under serum-free culture conditions. Stem Cell Res. Ther. 20150, 6, 235. [Google Scholar] [CrossRef] [PubMed]
- Darnell, M.; Gu, L.; Mooney, D. RNA-seq reveals diverse effects of substrate stiffness on mesenchymal stem cells. Biomaterials 2018, 181, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Abagnale, G.; Steger, M.; Nguyen, V.H.; Hersch, N.; Sechi, A.; Joussen, S.; Denecke, B.; Merkel, R.; Hoffmann, B.; Dreser, A. Surface topography enhances differentiation of mesenchymal stem cells towards osteogenic and adipogenic lineages. Biomaterials 2015, 61, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tan, K.; Zhou, Y.; Ye, Z.; Tan, W.-S. A combinatorial variation in surface chemistry and pore size of three-dimensional porous poly (ε-caprolactone) scaffolds modulates the behaviors of mesenchymal stem cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 59, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Lanniel, M.; Huq, E.; Allen, S.; Buttery, L.; Williams, P.M.; Alexander, M.R. Substrate induced differentiation of human mesenchymal stem cells on hydrogels with modified surface chemistry and controlled modulus. Soft Matter 2011, 7, 6501–6514. [Google Scholar] [CrossRef]
- Ejtehadifar, M.; Shamsasenjan, K.; Movassaghpour, A.; Akbarzadehlaleh, P.; Dehdilani, N.; Abbasi, P.; Molaeipour, Z.; Saleh, M. The effect of hypoxia on mesenchymal stem cell biology. Adv. Pharm. Bull. 2015, 5, 141. [Google Scholar] [CrossRef] [PubMed]
- Palomäki, S.; Pietilä, M.; Laitinen, S.; Pesälä, J.; Sormunen, R.; Lehenkari, P.; Koivunen, P. HIF-1α is upregulated in human mesenchymal stem cells. Stem Cells 2013, 31, 1902–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhu, Y.; Zhang, L.; Wu, T.; Wu, T.; Zhang, W.; Decker, A.M.; He, J.; Liu, J.; Wu, Y. Human Stem Cells Overexpressing miR-21 Promote Angiogenesis in Critical Limb Ischemia by Targeting CHIP to Enhance HIF-1α Activity. Stem Cells 2016, 34, 924–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Deng, Q.; Song, W.; Zhang, H.; Li, Y.; Yang, Y.; Fan, X.; Liu, M.; Shang, J.; Sun, C. MIF Plays a Key Role in Regulating Tissue-Specific Chondro-Osteogenic Differentiation Fate of Human Cartilage Endplate Stem Cells under Hypoxia. Stem Cell Rep. 2016, 7, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Pingguan-Murphy, B.; Abas, W.A.B.W.; Yong, K.W.; Poon, C.T.; Azmi, M.A.N.; Omar, S.Z.; Chua, K.H.; Xu, F.; Safwani, W.K.Z.W. In situ normoxia enhances survival and proliferation rate of human adipose tissue-derived stromal cells without increasing the risk of tumourigenesis. PLoS ONE 2015, 10, e0115034. [Google Scholar] [CrossRef] [PubMed]
- Tamama, K.; Kawasaki, H.; Kerpedjieva, S.S.; Guan, J.; Ganju, R.K.; Sen, C.K. Differential roles of hypoxia inducible factor subunits in multipotential stromal cells under hypoxic condition. J. Cell. Biochem. 2011, 112, 804–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramalingam, M.; Jabbari, E.; Ramakrishna, S.; Khademhosseini, A. Micro and Nanotechnologies in Engineering Stem Cells and Tissues; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Karageorgiou, V.; Kaplan, D. Porosity of 3D biomaterial scaffolds and osteogenesis. Biomaterials 2005, 26, 5474–5491. [Google Scholar] [CrossRef] [PubMed]
- Martin, Y.; Vermette, P. Bioreactors for tissue mass culture: Design, characterization, and recent advances. Biomaterials 2005, 26, 7481–7503. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kwiatkowska, B.; Lu, H.; Voglstätter, M.; Ueda, E.; Grunze, M.; Sleeman, J.; Levkin, P.A.; Nazarenko, I. Collaborative Action of Surface Chemistry and Topography in the Regulation of Mesenchymal and Epithelial Markers and the Shape of Cancer Cells. ACS Appl. Mater. Interfaces 2016, 8, 28554–28565. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhang, B.; Kasoju, N.; Ma, J.; Yang, A.; Cui, Z.; Wang, H.; Ye, H. Differential and Interactive Effects of Substrate Topography and Chemistry on Human Mesenchymal Stem Cell Gene Expression. Int. J. Mol. Sci. 2018, 19, 2344. https://doi.org/10.3390/ijms19082344
Li Q, Zhang B, Kasoju N, Ma J, Yang A, Cui Z, Wang H, Ye H. Differential and Interactive Effects of Substrate Topography and Chemistry on Human Mesenchymal Stem Cell Gene Expression. International Journal of Molecular Sciences. 2018; 19(8):2344. https://doi.org/10.3390/ijms19082344
Chicago/Turabian StyleLi, Qiongfang, Bo Zhang, Naresh Kasoju, Jinmin Ma, Aidong Yang, Zhanfeng Cui, Hui Wang, and Hua Ye. 2018. "Differential and Interactive Effects of Substrate Topography and Chemistry on Human Mesenchymal Stem Cell Gene Expression" International Journal of Molecular Sciences 19, no. 8: 2344. https://doi.org/10.3390/ijms19082344