Abstract
To identify circulating emerging/reemerging viral strains and epidemiological trends, an influenza sentinel surveillance network was established in Shandong Province, China, in 2005. Nasal and/or throat swabs from patients with influenza-like-illness were collected at sentinel hospitals. Influenza viruses were detected by reverse transcription polymerase chain reaction (RT-PCR) or virus isolation. From October 2005 to March 2012, 7763 (21.44 %) of 36,209 swab samples were positive for influenza viruses, including 5221 (67.25 %) influenza A and 2542 (32.75 %) influenza B. While the influenza viruses were detected year-round, their type/subtype distribution varied significantly. Peak influenza activity was observed from November to February. The proportion of laboratory-confirmed influenza cases was highest among participants aged 0-4 years (14.97 %) in the 2005-2009 and 2010-2012 influenza seasons and the positivity rate of influenza A(H1N1)pdm09 was highest in the 15 to 24 year age group during the 2009-2010 influenza season. Genetic analysis of hemagglutinin (HA) and neuraminidase (NA) genes revealed that the viruses matched seasonal influenza vaccine strains in general, with some amino acid mutations. Influenza A(H1N1)pdm09 strains isolated in Shandong Province were characterized by an S203T mutation that is specific to clade 7 isolates. This report illustrates that the Shandong Provincial influenza surveillance system was sensitive in detecting influenza virus variability by season and by genetic composition. This system will help official public health target interventions such as education programs and vaccines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Influenza viruses are important viral respiratory pathogens that can cause severe illness and death. Influenza A viruses have been responsible for several human pandemics in recent history. For example, H1N1 in 1918-20 caused an estimated 50 million excess deaths, H2N2 in 1957-8, 1 million deaths, H3N2 in 1968-9 up to 1 million deaths and H1N1pdm09 in 2009 an estimated 200,000 deaths. Each year influenza epidemics also occur in many parts of the world and are a cause of high mortality and morbidity [1]. The World Health Organization (WHO) estimates that, globally, there are 3–5 million severe cases and 250,000–500,000 deaths caused by influenza every year, with most deaths occurring among elderly populations [2]. Antigenic drift plays an important role in the occurrence of influenza epidemics [1]. Because of the high mutation rate of influenza viruses, year-round continuous monitoring for viral genetic change is essential.
Influenza surveillance is an important tool for monitoring circulating strains, detecting emerging/reemerging viruses, and identifying epidemiological trends in different geographical areas. It is also relevant for the evaluation of antigenic and genetic characteristics of circulating strains in comparison with recommended vaccine strains.
The Global Influenza Surveillance and Response System (GISRS) is active in 112 countries, which together comprise about 90 % of the global population. China is part of GISRS. Influenza surveillance in Shandong Province started in 2005 with three virology laboratories and seven sentinel hospitals. In response to the emergence of influenza A (H1N1) pdm09 in North America, the influenza surveillance network expanded to 18 virology laboratories and 27 sentinel hospitals in 2009, covering all districts in Shandong Province. Before 2009, the surveillance was conducted from October to March, when influenza was more common. Since May 2009, surveillance has been conducted year-round.
Shandong Province, located in eastern China (latitude 34°38’25” to 38°38’69”N), has a long coastline facing Japan and the Republic of Korea and an estimated population of 99.70 million. In this report, we summarize influenza surveillance data, analyze the influenza activity and the genetic features of virus strains in Shandong Province during the period from 2005 to 2012.
Materials and methods
Case definition of influenza-like illness (ILI) and the study population
A case of influenza-like illness (ILI) was defined as a person who presents with a history of sudden onset of fever ≥38 °C in the preceding 3 days, with cough or sore throat [3]. All specimens were collected from seven sentinel surveillance hospitals from October 2005 to March 2009 and 27 hospitals from May 2009 to March 2012 (Fig. 1).
Laboratory diagnosis
Nasal or throat swabs from ILI patients were collected and transported to the virology laboratory according to the National Technical Documents for Influenza published by the Chinese Center for Disease Control and Prevention (CCDC) [4]. From 2005 to March 2009, all samples from sentinel surveillance hospitals were tested for influenza viruses by virus isolation in MDCK cells. From May 2009, the clinical specimens were identified by viral culture and/or RT-PCR and/or real-time RT-PCR by the 18 virology laboratories in Shandong Province.
Virus isolation and identification
Nasal or throat swabs were used to inoculate Madin-Darby canine kidney (MDCK) cells. Upon the appearance of a cytopathic effect (75-100 %) or after 7 days of culture, the cell supernatant fluid was harvested and a hemagglutination (HA) test performed. If the HA titer was greater than or equal to 1:8, the sample was considered positive. If the HA titer was less than 1:8 or 0, the cell supernatant was subjected to an additional passage. For viruses with an HA titer ≥1:8, the influenza virus subtype was determined using the hemagglutination inhibition (HI) assays with the reference antisera distributed each year by the Chinese National Influenza Center (CNIC). For viruses with an HA titer <1:8, the subtype was identified by RT-PCR or real-time RT-PCR according to the technical documents published by CCDC [4]. All of the isolated viruses were sent to CNIC.
RNA extraction and RT-PCR detection
Viral RNA was extracted from nasal or throat swabs and viruses using a QIAGEN RNA Mini Kit (cat. no. 74104, Germany) according to the manufacture’s protocol. A one-step RT-PCR or a real-time RT-PCR was used to detect different influenza virus subtypes using QIAGEN kits. Samples were first classified as influenza A or influenza B. Most influenza-A-positive samples were further tested to identify different subtypes following Chinese technical documents [4].
Sequencing and phylogenetic analysis
To construct a phylogenetic tree, 143 isolated influenza viruses with high viral load from different surveillance years were selected: 47 influenza A(H3N2), 39 influenza B and 57 influenza A(H1N1)pdm09 viruses. The HA or NA gene was amplified using primers described previously [5–7]. The resulting single fragments were sent to Huada Gene Biotechnology Company (Beijing China) for sequencing by the dideoxynucleotide chain termination method. The gene sequences were aligned using the Clustal W program for the major coding regions: HA1 (966 bp) for A (H3N2), HA1 (1002 bp) for influenza B, HA (1550 bp) and NA (1250 bp) for influenza A (H1N1) pdm09. Neighbor-joining (NJ) phylogenetic trees were constructed using MEGA 5.0 software, using Kimura’s two-parameter distance model with 1000 bootstrap replicates. Bootstrap values above 90 were labeled on major tree branches for reference. All gene sequences in this study were deposited in the GeneBank database under accession numbers CY063666-063702, CY064922-064936, KR107136-107200, and KT424131-179.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) and Excel 2007. Proportions of influenza-positive tests in different age and sex groups were compared by chi-square test. A multivariate logistic regression model was applied to examine the relationship between the positive rate and the different age groups. P ≤ 0.05 was considered statistically significant.
Results
Influenza positivity in sentinel surveillance hospitals in Shandong Province
A total of 36,209 ILI samples were collected from 2005-12 and tested for influenza virus, by virus isolation, real-time RT-PCR or RT-PCR (since May 2009). Of these, 21.44 % (7763/36209) were positive for influenza viruses (Table 1). The highest prevalence of influenza was 39.72 % during the 2009-2010 surveillance season, followed by the 2007-2008 season (25.88 %). This increase was primarily due to the emergence of influenza A (H1N1) pdm09 in 2009 and changes in detection methods.
Influenza types and subtypes in Shandong Province
While the influenza viruses were detected year-round, the types/subtype varied considerably. The percentage distribution of influenza viruses by month was plotted over the total number of positive samples for influenza viruses in the respective year (Table 1 and Fig. 2). Based on the HI assay and the RT-PCR results, 67.25 % (5221/7763) of the positive samples were influenza A:263 seasonal A (H1N1), 3143 influenza A (H1N1) pdm09, 1153 seasonal A (H3N2) and 662 influenza A non-typed. The remaining positives were influenza B (32.75 %, 2542/7763). Influenza A was predominant during most of the surveillance years (ranging from 51 % to 95.35 %), except in the 2011-12 influenza season (20.58 %). There were at least two influenza virus subtypes co-circulating in Shandong Province in each surveillance year. The prevalent influenza viruses were seasonal A (H1N1) and seasonal A (H3N2) in the 2005-06 and 2006-07 influenza season, respectively. There was an equal distribution of seasonal A (H3N2) and influenza B during 2007-08 and an increase in prevalence of seasonal A (H1N1) during the 2008-09 influenza season. In 2009-10, seasonal A (H1N1) was completely replaced with influenza A (H1N1) pdm09 virus. The seasonal A (H3N2) viruses circulated at a low level in 2008-09, and their activity increased just before emergence of influenza A (H1N1) pdm09 in September 2009. In the 2010-11 influenza seasons, there was co-circulation of influenza B, seasonal A (H3N2) and influenza A (H1N1) pdm09 viruses. The influenza A (H1N1) pdm09 virus demonstrated a very discrete pattern with peaks in November, 2009 and January, 2011 (Fig. 2). In the 2011-12 influenza season, influenza B virus became the predominant virus in Shandong Province.
Seasonality
Figure 2 demonstrates the seasonality of influenza activity. In the four influenza seasons from 2005 to 2009, one peak was consistently observed from December to March, which is considered the winter season in Shandong Province. Samples from December and January accounted for 59.97 % of all positive samples during the four influenza seasons.
In the 2009-10 influenza season, a major peak was observed from September to December because of the pandemic caused by influenza A (H1N1) pdm09. With the decrease in influenza A (H1N1) pdm09, the peak and the positive rate sharply declined. In the 2010-12 influenza seasons, the greatest influenza activity was observed during January and February, with minor influenza A (H3N2) activity in September 2010.
Age and sex distribution
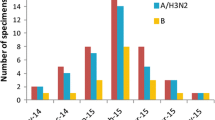
The median age of ILI patients enrolled in the study was 16.41 years (range: 1 month-95 years), and the mean age was 9 years. Among ILI cases, children under 5 years of age constituted 31.82 %. The under-5 age group also accounted for the highest percentage of laboratory-confirmed cases in the 2005-09 and 2009-12 influenza seasons, while the age group 15-24 years was the highest in the 2009-10 pandemic influenza season (Fig. 3).
Influenza-like illness (ILI) cases by age, 2005-2012. For the blue bar, the axis shows the percentage of subjects in the different age groups for the collected ILI samples (N = 24,323 in 2005-2012 except 2009-2010, N = 11,886 in 2009-2010). For the red bar, the axis shows the positive rate in the different age groups
There was a statistically significant difference in the proportion of influenza-virus-positive cases among different age groups (0-4, 5-14, 15-24, 25-59, ≥60 years old); (p < 0.0001). The frequency of influenza A was significantly lower among children (<15 years) than that among adults (5.64 % vs. 7.32 %, p < 0.0001), in the surveillance years 2005-12 except the 2009-10 season. For influenza B, the positive rate among children (<15 years) was statistically significantly higher (p < 0.0001, OR: 5.517, 95 % CI: 4.649-6.548) than that among adults during all these periods. In the 2009-2010 surveillance season, further analysis showed that influenza A(H1N1)pdm09 in the 15 to 24-year-old (OR 3.735, 95 % CI 2.697-5.172, P < 0.0001) and 5 to 14-year-old (OR 2.386, 95 % 1.722-3.306, P < 0.0001) groups were significantly higher compared with the ≥60-year-old group.
From 2005 to 2012, there was no significant difference in the influenza-positive proportion among males and females, and the male-to-female ratio was 1.
Phylogenetic analysis of influenza viruses
A/H3N2 influenza viruses
The HA gene of the 47 circulating influenza A (H3N2) viruses identified in Shandong Province during the 2006-11 surveillance seasons were compared with the vaccine strains. As seen in Figure 4, H3N2 Shandong strains grouped into three clusters: the 2006-2008 isolates, with 99.1 %-100.0 % similarity clustered with the A/Brisbane/10/07-like strain, which was a 2007-2009 northern hemisphere vaccine component. In 2008-2009 a change (K173Q/E) in the circulating strain took place. In the 2009-2011 influenza season, influenza A(H3N2) virus with 96.9 %-100.0 % similarity, fell into three phylogenetic clades. Some isolates still grouped with A/Brisbane/10/07, while most strains fell into the clades represented by A/Perth/16/09 and A/Victoria/208/2009, with the amino acid mutation T212A. The nine potential N-glycosylation sites were found to be conserved in all isolates during the 2006-2011 seasons.
Influenza A (H1N1) pdm09 virus
A phylogenetic tree of the HA and NA nucleotide sequences of the Shandong Province strains from the 2009-2011 seasons and the vaccine are shown in Figure 5 and Figure 6. Among the Shandong influenza A (H1N1) pdm09 strains during 2009-2011, the gene sequence similarities in HA and NA were 98.4 %-100 % and 98.5 %-100 %, respectively. All isolates were descended from A/California/2009-like viruses. The HA phylogeny shows that the amino acid substitutions P83S, S203 and I321V were found in all of the Shandong isolates. The substitutions S128P and D86G/Y were identified in 50 % and 30 % of the A/H1N1 pdm09 viruses in the 2009-2010 and 2010-2011 season, respectively. The selected viruses had histidine (H) at position 275 in the NA gene, indicating sensitivity to neuraminidase inhibitors. Other substitutions, V106I and N248D in the NA gene, were present in the analyzed viruses.
Phylogenetic tree comparing the HA genes of A/H1N1pdm09 Shandong strains and the vaccine strain. The representative strains from A/California/07/2009 (H1N1) (red triangle) and Shandong (red circle) and other parts of the world were used to generate the phylogenetic tree. A/Sichuan/01/2009 (red square) was the first influenza A(H1N1)pdm09 strain in China, A/Shandong/01/2009 (red triangle) was the first influenza A(H1N1)pdm09 strain in Shandong Province. The characteristic amino acid (S203T) is shown at the root of the branch
Influenza B virus
Phylogenetic analysis based on the HA1 sequences of 39 influenza B virus isolates from 2006-2012 revealed that influenza B virus was present as a mixed population of viral variants, since viruses belonging to both B/Victoria and B/Yamagata lineages co-circulated in different proportions (Fig. 7). The Shandong strains from 2006-2012 grouped into four clades, represented by the corresponding vaccine strains. The substitutions D197N, and S230D/G and 163 deletions were observed in all Shandong B/Yamagata isolates, and the B/Victoria isolates also had L58P and N75K substitutions, with the exception of four strains (Fig. 7).
Discussion
In this report, we have described the influenza virus seasonality and evolutional analysis of influenza virus in Shandong Province during the period from 2005 to 2012. We found that influenza viruses affected all age groups and circulated seasonally during the winter months of November through February, with peak circulation occurring in December and January, similar to patterns documented in the temperate regions of the northern hemisphere [8–12]. The epidemiological data revealed that multiple influenza subtypes co-circulated in Shandong Province and that the predominant subtype varied each year (Table 1 and Fig. 2), which may partly be due to limited immunological cross-reactivity between influenza subtypes. There was also evidence that the dynamic patterns and displacements of different subtypes were linked to host factors, environmental factors, and the virulence and/or pathogenicity of each strain [13–15]. Genetic and phylogenetic analysis of influenza virus circulating in Shandong Province confirmed that the genetic makeup of influenza viruses may change from year to year.
A strong surveillance system that covered all of the diverse settings in Shandong Province enabled us to monitor the emergence of influenza A (H1N1) pdm09 virus throughout the province. After the detection of the influenza A (H1N1) pdm09 virus outbreak in North America in the spring of 2009, we observed a high prevalence of seasonal A/H3N2 in Shandong province, which was consistent with the other provinces in China [16]. By contrast, the United States reported that >90 % of the positive samples contained influenza A (H1N1) pdm09 virus during the first wave in 2009 [17]. To delay community outbreaks of influenza A (H1N1)pdm09 in China, a strict prevention strategy was implemented in the early stage of the pandemic, including border entry screening [18], resulting in only sporadic and imported cases. With the opening of school and the modulated prevention strategy in September 2009, the number of influenza A (H1N1) pdm09 patients gradually increased [19]. In October 2009, influenza A (H1N1) pdm09 became the predominant virus, and a distinct peak occurred in November 2009, and followed by a sharp decline in January 2010, when influenza B viruses became more prevalent. These observations are in agreement with those from other parts of the world, where influenza A (H1N1) pdm09 either completely replaced seasonal influenza or co-circulated with seasonal influenza viruses [20, 21]. At the same time, in response to the influenza A(H1N1)pdm09 pandemic, a free vaccination program was implemented in Shandong Province from October 2009 to March 2010, covering about 5 million people of all ages (data unpublished): 0-5 years (1.99 % of those who were vaccinated), 6-15 years (30.87 %), 16-24 years (27.4 %), 25-59 years (35.26 %) and ≥ 60 years (4.48 %). This helped prevent the virus from circulating more widely among the population. At the same time, we found that the first wave of the 2009 pandemic started to decline in December 2009, which was in agreement with the national data in China [18]. The decline was attributed to protection after natural infections or vaccination during these periods.
In the 2010-2011 surveillance season, influenza B virus circulation was observed in the early phase, then influenza A/H3N2 in August-November 2010, followed by a second wave of influenza A (H1N1) pdm09 in January 2011, similar to what is observed presently in most countries or districts around the globe [22–24]. The “second-wave” pattern that was observed could also be seen in the 1918-1919 H1N1 pandemic and 1957-1958 H2N2 pandemic, but only a single peak was observed in the 1968 H3N2 pandemic, which demonstrates the unpredictable nature of influenza pandemics [25]. We postulate that at least four factors play key roles in this phenomenon. First, the antibody against influenza A (H1N1) pdm09 virus acquired by natural infections or vaccination or prior immunity may have decreased. Wang et al. reported that at least 50 % of vaccinees and recovered patients lost protective immunity after 6 months [26]. Dorigatti et al. [27] stated that the main reason for the 2010-2011 influenza A (H1N1) pdm09 wave was not a decay of homologous immunity but decreased levels of the preexisting protective cross-reactive immunity in adults during 2009 by the winter of 2010 in the United Kingdom, which resulted in more susceptible people. In Shandong Province, the result of the serological surveys in 2010 showed that the seroprevalence rate in September was 25.85 %, the positive rate of vaccinated people decreased from 50.9 % to 25.43 %, and the positive rate of the age group ≥60 years increased from 8.4 % to 28.43 % from January to September 2010 [28, 29]. Thus, the decay of homologous and/or heterologous immunity may have resulted in the second wave of the influenza A (H1N1) pdm09 epidemic in the 2010-2011 surveillance seasons in Shandong Province. Second, the transmissibility of influenza A (H1N1) pdm09 virus may have increased, partly because of the genetic adaptation of the virus to the human host. Dorigatti et al. also showed that viral transmissibility increased in England [27], with the effective reproduction number Re increasing from 1.2 in the second wave to 1.5 by the start of the third wave (Fall 2010). Third, the shift in the age distribution of cases between the two waves contributed to the appearance of the second wave [23, 30]. Compared with the 2009-2010 epidemics, for the age group ≥25 years, the percent of the verified cases in all positive cases increased from 25.63 % to 40.4 %, while it decreased from 69.29 % to 38.2 % for the age group of 5-24 years in the second wave (data unpublished). Lastly, some environmental factors may have contributed to the second wave. There is evidence that drastic changes in meteorological parameters, such as humidity and cold weather [31, 32], can modulate viral survival and transmissibility and also affect human immunity. According to the Shandong Weather Bureau, the average temperature in January 2011 was 2 degrees centigrade lower than that in January 2010, and the average amount of the rain decreased 5 mm, which may have substantially affected the ability of the virus to be transmitted among the population.
Age-group analysis revealed that the highest rate of influenza virus positivity except for the 2009-2010 influenza season was observed among children in the age group of 0-4 years, which could be due to timely medical diagnosis and treatment when the children were ill, compared with young adults and adults, who prefer to remain at home and use self-administered medication, which was also verified by the reported ILI cases from the sentinel hospitals [33, 34]. At the same time, the highest rate of influenza A (H1N1) pdm09 positivity in the 2009-2010 season was in the age group of 15-24 years, followed by 5-14 years, which is consistent with reports from Mongolia (16-24 years), Taiwan (5-19 years) and Delhi (5-18 years and 18-25 years). These may be explained by their behavior and the high exposure rates among school age-children [35, 36].
Diversity in circulating influenza types and subtypes poses a real challenge to vaccine strategies [37]. The viral antigenic and evolutional analysis of circulating influenza viruses is considered the best method to recommend the most suitable influenza vaccines for the northern and southern hemisphere. Phylogenetic analysis of circulating viruses showed a general good match between recommended vaccine strains and circulating viruses in the 2006-2012 surveillance seasons, whereas some strains were more divergent in different surveillance seasons, such as influenza A (H3N2). In the 2007-2008 and 2008-2009 seasons, most of the Shandong influenza A(H3N2) viruses had high similarity to the vaccine strain A/Brisbane/10/2007, while some strains gradually evolved with the signature genetic marker K173Q/E after Spring, 2008. In the 2009-2010 season, the prevalent strains were replaced by an A/Perth/16/2009-like strain, an A/Victoria/208/2009-like strains evolved into the predominant strains until 2011, with the main amino acid mutation T212A. The strains fell into two clades in 2009-2011. At the same time, the amino acids in the H3 HA1 from 2006 to 2011 had a total of 50 amino acid substitutions, 14 of which were located at well-known antigenic sites.
Like other circulating strains in China, the influenza (A/H1N1) pdm09 virus isolates were characterized by three fixed mutations, P83S, S203T (except Chinese reference stain A/Sichuan/SWL01/2009) and I321V, with respect to A/California/07/2009. All Shandong influenza A (H1N1) pdm09 strains analyzed belonged to clade 7 with the signature sequence S203T in the HA1 protein [38]. Certain amino acid substitutions such as D222G in HA have been reported in connection with severe disease and poor outcome [39, 40], but none of Shandong isolates analyzed in this study had this substitution. The significance merits further study, especially in more-serious cases reported from Shandong province. Only one isolate, A/Shandong-Lixia/SWL131/2011, had the R257K substitutions reported previously in Finland [41]. The significance of substitution at this position for disease severity has not been clearly demonstrated. In the NA gene, the clade-7-specific substitutions (V106I and N248D) were uniformly seen in the analyzed viruses, while the characteristic Y275H substitutions reportedly associated with drug resistance were absent.
In conclusion, our study clearly illustrates that influenza was an important cause of ILI. Analysis of the epidemiological and virological data, together with the genetic and antigenic data, has also provided insights into the pattern of virus spread, the genetic diversity during seasonal epidemics, and the dynamics of evolution. Continuous surveillance of the dynamics of influenza virus subtypes and genetic change throughout consecutive years is necessary to find new antigenic or drug-resistant variants. This will further facilitate pandemic planning and response capacity at the national as well as the global level.
References
Shih SR, Chen GW, Yang CC, Yang WZ, Liu DP, Lin JH et al (2005) Laboratory-based surveillance and molecular epidemiology of influenza virus in Taiwan 23(4):1651–1661
World Health Organization (WHO) (2014) Fact sheet 211: influenza (seasonal). http://www.who.int/mediacentre/factsheets/fs211/en/. Accessed Mar 2014
National Health and Family Planning Commission of the PRC (2015) National influenza surveillance program. http://www.nhfpc.gov.cn/jkj/s3577/201009/3fa356d0f4834d408fde6c12891a6482.shtml,2010. Accessed 24 June 2015
Chinese Center for disease control and prevention (2011) National technical documents for influenza 2011. http://www.cnic.org.cn/chn/down/showdown.php?downid=663. Accessed 24 June 2015
Shu YL, Zhang Y, Wen LY, Li Z, Guo JF, Wang M et al (2005) Antigenic and genetic study of hemagglutinin gene of influenza virus (H3N2) circulated in China in 2004. Chinese J Exp Clin Virol 19(4):362–365 (In Chinese)
Zhang Y, Wen LY, Zhao X, Li Z, Guo JF, Xu CL et al (2006) Antigenic and genetic study of influenza virus B circulated in China in 2004–2005. Chinese J Exp Clin Virol 20(2):11–13 (In Chinese)
Liu T, Lin Y, Zhang SY, Wang S, Wang YL, Yin YY et al (2011) Etiological and molecular characteristics of pandemic influenza A (H1N1) virus during 2009–2010 in Shandong Province. Chinese J Virol 27(3):224–230 (In Chinese)
Cox NJ, Subbarao K (2000) Global epidemiology of influenza: past and present. Annu Rev Med 51:407–421
Li C, Freedman M (2009) Seasonal influenza: an overview. J Sch Nurs 25:4S–12S
Jennings LC (1999) Influenza surveillance in New Zealand. Vaccine 17:S115–S117
Zambon MC, Stockton JD, Clewley JP, Fleming DM (2001) Contribution of influenza and respiratory syncytial virus to community cases of influenza-like illness: an observational study. Lancet 27:1410–1416
Barakat A, Ihazmad H, Benkaroum S, Cherkaoui I, Benmamoun A, Youbi M, El Aouad R (2011) Influenza surveillance among outpatients and inpatients in Morocco, 1996–2009. PLoS One 6(9):e24579
World Health Organization (2011) WHO global influenza surveillance network: manual for the laboratory diagnosis and virological surveillance of influenza
Tamerius J, Nelson MI, Zhou SZ, Viboud C, Miller MA, Alonso WJ (2011) Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ Health Perspect 119(4):439–445
Tscherne DM, Garcia-Sastre A (2011) Virulence determinants of pandemic influenza viruses. J Clin Invest 121(1):6–13
Chinese National Influenza Center (2009) Weekly influenza surveillance (36 week, 2009). http://www.chinaivdc.cn/cnic/zyzx/lgzb/201605/t20160521_129750.htm. Accessed Sep 2009 (In Chinese)
Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team; Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S et al (2009) Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine origin influenza A (H1N1) virus in humans. N Engl J Med 360:2605–2615
Yu HJ, Cauchemez S, Donnelly CA, Zhou L, Feng LZ et al (2012) Transmission dynamics, border entry screening, and school holidays during the 2009 influenza A (H1N1) pandemic, China. Emerg Infect Dis 18(5):758–766
Chao DL, Halloran ME, Longini IM Jr (2010) School opening dates predict pandemic influenza A(H1N1) outbreak in the United States. JID 202(6):877–880
Tang JWT, Lee CK, Lee HK, Loh TP, Chiu L, Tambyah PA (2010) Tracking the emergence of pandemic influenza A/H1N1/2009 and its interaction with seasonal influenza viruses in Singapore. Ann Acad Med Singap 39:291–294
Mathews JD, Chesson JM, McCaw JM, McVernon J (2009) Understanding influenza transmission, immunity and pandemic threats. Influ Other Respir Virus 3:143–149
Athanasiou M, Baka A, Andreopoulou A, Spala G, Karageorgou K, Kostopoulos L et al (2011) Influenza surveillance during the post-pandemic influenza 2010/11 season in Greece, 04 October 2010 to 22 May 2011 Eur Surveill 16(44)
Chuang J-H, Huang AS, Huang W-T, Liu M-T, Chou J-H, Chang FY et al (2012) Nationwide surveillance of influenza during the pandemic (2009–10) and post-pandemic (2010–11) periods in Taiwan. PLoS One 7(4):e36120
Wangchuk S, Thapa B, Zangmo S, Jarman RG, Bhoomiboonchoo P, Gibbons RV (2013) Influenza surveillance from November 2008 to 2011; including pandemic influenza A(H1N1)pdm09 in Bhutan. Influ Other Respir Virus 7(3):426–430
Jhung Michael A, Swerdlow David, Olsen Sonja J, Jernigan Daniel, Biggerstaff Matthew, Kamimoto Laurie et al (2011) Epidemiology of 2009 pandemic influenza A (H1N1) in the United States. Clin Infect Dis 52(S1):S13–S26
Wang M, Yuan J, Li T, Liu Y, Wu J, Di B et al (2011) Antibody dynamics of 2009 influenza A (H1N1) virus in infected patients and vaccinated people in China. PloS One 6:e16809
Dorigatti Ilaria, Cauchemez Simon, Ferguson Neil M (2013) Increased transmissibility explains the third wave of infection by the 2009 H1N1 pandemic virus in England. PNAS 110(33):13422–13427
Liu Ti, Li Zhong, Song Shaoxia, Zhang Shengyang, Wang Yulu, Lin Yi et al (2012) Post-pandemic seroprevalence of 2009 pandemic influenza A (H1N1) virus in Shandong Province, China. Jpn J Infect Dis 65:410–414
Zhang SY, Lin Y, Wang S, Ren YY, Liu T, Wang YL et al (2012) Serological survey of pandemic influenza A(H1N1) antibodies among healthy individuals in Shandong Province, in 2010. J Pathog Biol 7(1):10–12 (In Chinese)
Merler Stefano, Ajelli Marco, Pugliese Andrea, Ferguson Neil M (2011) Determinants of the spatiotemporal dynamics of the 2009 H1N1 pandemic in Europe: implications for real-time modeling. PLOS Comput Biol 7(9):e1002205
Shaman J, Kohn M (2009) Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci USA 106:3243–3248
Steel J, Palese P, Lowen AC (2011) Transmission of a 2009 pandemic influenza virus shows a sensitivity to temperature and humidity similar to that of an H3N2 seasonal strain. J Virol 85:1400–1402
Orelle Arnaud, Razanajatovo Norosoa Harline, Rajatonirina Soatiana, Hoffmann Jonathan, Randrianasolo Laurence, Razafitrimo Girard Marcellin et al (2012) Epidemiological and virological characterization of 2009 pandemic influenza A virus subtype H1N1 in Madagascar. J Infect Dis 206(S1):S140–S147
Chinese National Influenza Center (2010) Weekly influenza surveillance (12 week, 2010). http://www.chinaivdc.cn/cnic/zyzx/lgzb/201605/t20160521_129777.htm. Accessed Apr 2010 (In Chinese)
Devaux I, Kreidl P, Penttinen P, Salminen M, Zucs P, Ammon A (2010) Initial surveillance of 2009 influenza A(H1N1) pandemic in the European union and European economic area, April–September 2009. Euro Surveill 15:19740
Baguelin M, Hoschler K, Stanford E et al (2011) Age-specific incidence of A/H1N1 2009 influenza infection in England from sequential antibody prevalence data using likelihood-based estimation. PLoS One 6:e17074
Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A et al (2009) Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 325:197–201
Potdar VA, Chadha MS, Jadav SM, Mullick J, Cherian SS, Mishra AC et al (2010) Genetic characterization of the influenza a pandemic (H1N1) 2009 virus isolates from India. 2010. PloS One 5:e9693
Kilander A, Rykkvin R, Dudman SG, Hungnes O (2010) Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009–2010. Euro Surveill 15:19498
Mak GC, Au KW, Tai LS, Chuang KC, Cheng KC, Shiu TC et al (2010) Association of D222G substitution in haemagglutonin of 2009 pandemic influenza A (H1N1) with severe disease. Euro Surveill 15:19534
Ikonen N, Haanpää M, Rönkkö E, Lyytikäinen O, Kuusi M, Ruutu P et al (2010) Genetic diversity of the 2009 pandemic influenza A (H1N1) viruses in Finland. PLoS One 5(10):e13329
Acknowledgments
The authors acknowledge the effort and contribution of the surveillance physicians of the sentinel hospitals and labs in Shandong Province, Xiyan Xu and Shang Mei (Centers for Disease Control and Prevention, United States) and Gregory C. Gray (Duke University) for the manuscript management.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The study was approved by the Ethics Committee of the Shandong Center for Disease Control and Prevention. Informed written consent was obtained from all subjects (or their participants’ guardians) who had a nasal or throat swab specimen collected.
Funding
This work was supported by National Science and Technology Major Project (2014ZX09509001) and Scientific and Technological Projects of Shandong Province (2009GG10002054).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Z. Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, T., Li, Z., Lin, Y. et al. Dynamic patterns of circulating influenza virus from 2005 to 2012 in Shandong Province, China. Arch Virol 161, 3047–3059 (2016). https://doi.org/10.1007/s00705-016-2997-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2997-2