Abstract
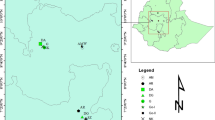
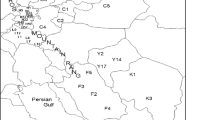
There are many valuable Tadehagi accessions in southwest China, but it is unknown that the genetic diversity and phylogenetic relationship of these Tadehagi resources. This report is the first study in which 41 primers of inter-simple sequence repeat (ISSR) were used to assess the genetic diversity of 36 Tadehagi accessions from 3 provinces in the southwest of China. Totally, 30 usable ISSR primers detected 163 polymorphic bands among the 36 accessions, which suggested high utility of ISSR primers in the genetic analysis of Tadehagi accessions. Genetic similarity coefficients among all of the accessions ranged from 0.54 to 0.92 with the average of 0.79 based on the ISSR data, indicating high level of genetic variation in Tadehagi resources from the southwest of China. As for the 3 population, Hainan population had the maximum average genetic similarity coefficients of 0.81, while similarity coefficient of Guangxi and Yunnan population was 0.75 and 0.74, respectively. All the 36 Tadehagi accessions were divided into 4 groups in the UPGMA dendrogram constructed from genetic similarity coefficients. The Tadehagi accessions from Yunnan and Guangxi provinces showed more genetic variation and occupied the bottom of the dendrogram. On the contrary, those from Hainan Province had less genetic variation and clustered in the middle and top of the dendrogram. The information on the genetic diversity and phylogenetic relationship from this study is propitious to construct a core germplasm collection and develop novel Tadehagi cultivars with desired economic traits.



Similar content being viewed by others
References
Aphalo PJ, Ballaré CL (1995) On the importance of information-acquiring systems in plant-plant interactions. Funct Ecol 9:5–14
Ballaré CL, Scopel AL (1997) Phytochrome signalling in plant canopies: testing its population-level implications with photo-receptor mutants of Arabidopsis. Funct Ecol 11:441–450
Blair MW, Panaud O, McCouch SR (1999) Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.). Theor Appl Genet 98:780–792
Byrne M, Elliott CP, Yates CJ, Coates DJ (2008) Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conserv Genet 9(1):97–105
Ding G, Li X, Ding X, Qian L (2009) Genetic diversity across natural populations of Dendrobium officinale, the endangered medicinal herb endemic to China, revealed by ISSR and RAPD markers. Russ J Genet 45:327–334
Excoffier L, Laval G, Schneider S (2005) Arlequin: an integrated software package for population genetics data analysis, version 3.0. Evol Bioinform Online 1: 47–50
Heider B, Fischer E, Berndl T, Schultze-Kraft R (2009) Genetic relationships among accessions of four species of Desmodium and allied genera (Dendrolobium triangulare, Desmodium gangeticum, Desmodium heterocarpon, and Tadehagi triquetrum). Trop Conserv Sci 2:52–69
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS (2000) Genetic diversity and phylogenetic relationship as revealed by inter-simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Kalisz S, Hanzawa FM, Tonsor SJ, Thiede DA, Voigt S (1999) Ant-mediated seed dispersal alters pattern of relatedness in a population of Trillium grandiflorum. Ecology 80:2620–2634
Liu DQ, Guo XP, Lin ZX, Nie YC, Zhang XL (2006) Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol 53:1145–1152
Nagaoka T, Ogihara Y (1997) Applicability of inter-simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor Appl Genet 94:597–602
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3:93–114
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Reddy PM, Sarla N, Siddiq EA (2002) Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128:9–17
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, Setauket, New York
Shu AP, Kim JH, Zhang SY, Cao GL, Nan ZH, Lee KS, Lu Q, Han LZ (2009) Analysis on genetic similarity of japonica rice variety from different origins of geography in the world. Agric Sci China 8:513–520
Smith H (1990) Phytochrome action at high photon fluence rates: rapid extension rate responses of light-grown mustard to variations influence rate and red: far-red ratio. Photochem Photobiol 52:131–142
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 28:1409–1438
Thomas D, Schultze-Kraft R (1990) Evaluation of five shrubby legumes in comparison with Centrosema acutifolium, Carimagua, Colombia. Tropical Grasslands 24:87–92
Volis S, Yakubov B, Shulgina I, Ward D, Zur V, Mendlinger S (2001) Tests for adaptive RAPD variation in population genetic structure of wild barley, Hordeum spontaneum Koch. Biol J Linn Soc 74:289–303
Wen CS, Hsiao JY (1999) Genetic differentiation of Lilium longiflorum Thunb. var. scabrum Masam. (Liliaceae) in Taiwan using random amplified polymorphic DNAs and morphological characters. Bot Bull Acad Sinica (Taipei) 40:65–71
Wen DX, Lu MY, Tang RJ, Zheng XZ, Inoue K (2000) Studies on the chemical constituents of triquetrous tadehagi (Tadehagi triquetrum) (II). Chin Tradit Herbal Drugs 31:3–5
Xia T, Chen SL, Chen SY, Ge XJ (2005) Genetic variation within and among populations of Rhodiola alsia (Crassulaceae) native to the Tibetan Plateau as detected by ISSR markers. Biochem Genet 43:87–101
Xiang W, Li RT, Mao YL, Zhang HJ, Li SH, Song QS, Sun HD (2005) Four new prenylated isoflavonoids in Tadehagi triquetrum. J Agric Food Chem 53:267–271
Yang Y, Huang P (1995) Tadehagi Ohashi. In: Yang Y, Huang P, Fu P, Li J, Chen Y, Lee S, Chang B, Wei Y, Huang D, Wei C, Wu T, Wei S (eds) Flora of China, vol 41. Science Press, Beijing, pp 62–65
Yue MF, Zhou RC, Huang YL, Xin GR, Shi SH, Feng L (2010) Genetic diversity and geographical differentiation of Desmodium triflorum (L.) DC. in south China revealed by AFLP markers. J Plant Biol 53:165–171
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
Financial support of this research from the Yunnan Provincial Natural Science Foundation (2008ZC036 M) was appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Diqiu Liu and Xin He contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, D., He, X., Liu, G. et al. Genetic diversity and phylogenetic relationship of Tadehagi in southwest China evaluated by inter-simple sequence repeat (ISSR). Genet Resour Crop Evol 58, 679–688 (2011). https://doi.org/10.1007/s10722-010-9611-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-010-9611-3