Abstract
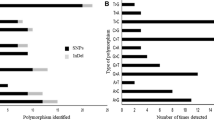
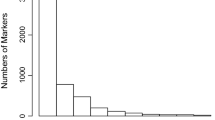
Coffee is one of the most widely consumed beverages and represents a multibillion-dollar global industry. Accurate identification of coffee cultivars is essential for efficient management, exchange, and use of coffee genetic resources. To date, a universal platform that can allow data comparison across different laboratories and genotyping platforms has not been developed by the coffee research community. Using expressed sequence tags (EST) of Coffea arabica, C. canephora and C. racemosa from public databases, we developed 7538 single nucleotide polymorphism (SNP) markers and selected 180 for validation using 25 C. arabica and C. canephora accessions from Puerto Rico. Based on the validation result, we designated a panel of 55 SNP markers that are polymorphic across the two species. The average minor allele frequency and information index of this SNP panel are 0.281 and 0.690, respectively. This panel enabled the differentiation of all tested accessions of C. canephora, which accounts for 79.2 % of the total polymorphism in the samples. Only 21.8 % of the polymorphic SNPs were detected in the 12 C. arabica cultivars, which, nonetheless, were able to unambiguously differentiate the 12 Arabica cultivars into ten unique genotypes, including two synonymous groups. Several local Puerto Rican cultivars with partial Timor pedigree, including Limaní, Frontón, and TARS 18087, showed substantial genetic difference from the other common Arabica cultivars, such as Catuai, Borbón, and Mundo Nuevo. This coffee SNP panel provides robust and universally comparable DNA fingerprints, thus can serve as a genotyping tool to assist coffee germplasm management, propagation of planting material, and coffee cultivar authentication.


Similar content being viewed by others
Abbreviations
- cDNA:
-
Complementary DNA
- DNA:
-
Deoxyribonucleic acid
- EST:
-
Expressed sequence tag
- PCR:
-
Polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
- USDA ARS:
-
United States Department of Agriculture, Agricultural Research Service
References
Afroz A, Khan MR, Komatsu S (2010) Determination of proteins induced in response to jasmonic acid and salicylic acid in resistant and susceptible cultivars of tomato. Protein Pept Lett 17:1–11
Anthony F, Astorga C, Avendaño J et al (2007) Conservation of coffee genetic resources in the CATIE field genebank. In: Engelmann F, Dulloo ME, Astorga C et al (eds) Conserving coffee genetic resources. Bioversity International, Rome, pp 23–34
Anthony F, Bertrand B, Quiros O et al (2001) Genetic diversity of wild coffee (Coffea arabica L.) using molecular markers. Euphytica 118:53–65
Combes MC, Dereeper A, Severac D et al (2013) Contribution of subgenomes to the transcriptome and their intertwined regulation in the allopolyploid Coffea arabica grown at contrasted temperatures. New Phytol 200:251–260
Cubry P, Musoli P, Legnaté H et al (2008) Diversity in coffee assessed with SSR markers: structure of the genus Coffea and perspectives for breeding. Genome 2008(51):50–63
Davis AP (2010) Six species of Psilanthus transferred to Coffea (Coffeeae, Rubiaceae). Phytotaxa 10:41–45
Davis AP (2011) Psilanthus mannii, the type species of Psilanthus, transferred to Coffea. Nordic J Bot 29:471–472
Davis AP, Govaerts R, Bridson DM et al (2006) An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot J Linn Soc 152:465–512
de Kochko A, Akaffou S, Andrade AC et al (2010) Advances in Coffea genomics. Adv Bot Res 53:23–63
Denoeud F, Carretero-Paulet L, Dereeper A et al (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184
Dereeper A, Bocs S, Rouard M et al (2015) The coffee genome hub: a resource for coffee genomes. Nucleic Acids Res 43:D1028–D1035
Dieringer D, Schlötterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Fang W, Meinhardt LW, Tan H et al (2014) Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers. Hortic Res 1:14035
FAOSTAT (2014) Food and Agriculture Organization of the United Nations, Statistics Division. Available online at: http://faostat.fao.org/
Felsenstein J (1989) Mathematics vs. evolution: mathematical evolutionary theory. Science 246:941–942
Fluidigm (2011) Fluidigm SNP Genotyping User Guide Rev H1, PN 68000098. South San Francisco, CA: Fluidigm Corporation. http://www.mscience.com.au/upload/pages/fluidigmtech/fluidigm-snp-genotyping-user-guide-151112.pdf
Geleta M, Herrera I, Monzón A et al (2012) Genetic diversity of Arabica coffee (Coffea arabica L.) in Nicaragua as estimated by simple sequence repeat markers. Sci World J 2012:939820
Gole TW, Denich M, Teketay D, Vlek PLG (2002) Human impacts on the Coffea arabica genepool in Ethiopia and the need for its in situ conservation. In: Engels JMM, Ramanatha Rao V, Brown AHD, Jackson MT (eds) Managing plant genetic diversity. CABI Publishing, Oxon, pp 237–247
Gross GG (1980) The biochemistry of lignification. Adv Bot Res 8:26–63
Hamon P, Hamon S, Razafinarivo NJ et al (2015) Coffea genome organization and evolution. In: Preedy VR (ed) Coffee in health and disease prevention. Academic, San Diego, pp 29–37
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
International Coffee Organization (2014) World coffee trade (1963–2013): a review of the markets, challenges and opportunities facing the sector. International Coffee Organization, London, ICC 111–5 Rev.1.29
Ji K, Zhang D, Motilal L et al (2013) Genetic diversity and parentage in farmer varieties of cacao (Theobroma cacao L.) from Honduras and Nicaragua as revealed by single nucleotide polymorphism (SNP) markers. Genet Resour Crop Evol 60:441–453
Krishnan S, Ranker TA (2012) Coffee genomics. In: Benkeblia N (ed) OMICs technologies: tools for food science. CRC Press, Boca Raton, pp 227–247
Lashermes P, Paczek V, Trouslot P et al (2000) Single-locus inheritance in the allotetraploid Coffea arabica L. and interspecific hybrid C. arabica x C. canephora. J Hered 91:81–85
Leroy T, De Bellis F, Legnate H, Musoli P, Kalonji A, Loor Solorzano RG, Cubry P (2014) Developing core collections to optimize the management and the exploitation of diversity of the coffee Coffea canephora. Genetica 142:185–99
Lewin B, Giovannucci D, Varangis P (2004) Coffee markets: new paradigms in global supply and demand. The International Bank for Reconstruction and Development, Agriculture and Rural Development Discussion Paper 3, 150 pp
Liu W, Xiao ZD, Bao XL et al (2015) Identifying litchi (Litchi chinensis Sonn.) cultivars and their genetic relationships using single nucleotide polymorphism (SNP) markers. PLoS ONE 10(8), e0135390
López-Gartner G, Cortina H, McCouch SR et al (2009) Analysis of genetic structure in a sample of coffee (Coffea arabica L.) using fluorescent SSR markers. Tree Genet Genomes 5:435–446
Macherel D, Lebrun, Gagnon M, Neuburger M, Douce R (1990) cDNA cloning, primary structure and gene expression for H-protein, a component of the glycine-cleavage system (glycine decarboxylase) of pea (Pisum sativum) leaf mitochondria. Biochem J 268:783–789
Martinelli F, Tonutti P (2012) Flavonoid metabolism and gene expression in developing olive (Olea europaea L.) fruit. Plant Biosyst - Int J Dealing Asp Plant Biol 146(sup1):164–170. doi:10.1080/11263504.2012.681320
Masumbuko LI, Bryngelsson T (2006) Inter simple sequence repeat (ISSR) analysis of diploid coffee species and cultivated Coffea arabica L. from Tanzania. Genet Resour Crop Evol 53:357–366
McClelland TB (1924) Coffee varieties in Porto Rico. Porto Rico Agricultural Experiment Station, Mayaguez, Porto Rico, Bulletin No. 30, 27 pp
Missio RF, Caixeta ET, Zambolim EM et al (2010) Polymorphic information content of SSR markers for Coffea spp. Crop Breed Appl Biotechnol 10:89–94
Moncada P, McCouch S (2004) Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome 47:501–509
Moncada P, Tovar E, Montoya JC, González A, Spindel J, McCouch S (2016) A genetic linkage map of coffee (Coffea arabica L.) and QTL for yield, plant height, and bean size. Tree Genet Genomes 12:5
Mondego JMC, Vidal RO, Carazzolle MF et al (2011) An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and Coffea canephora. BMC Plant Biol 11:30
Monroig Inglés MF (n. d.) Descripción de variedades de Coffea arabica más cultivadas en Puerto Rico. Available online: http://academic.uprm.edu/mmonroig/id45.htm
Moore PH, Ming R (eds) (2008) Genomics of tropical crops plants. Springer Science + Business Media, New York, 581 pp
Nijveen H, van Kaauwen M, Esselink DG et al (2013) QualitySNPng: a user-friendly SNP detection and visualization tool. Nucleic Acids Res 41:W587–W590
Osorio N (2002) The global coffee crisis: a threat to sustainable development. International Coffee Organization, London. Available online: http://www.ico.org/documents/globalcrisise.pdf
Peakall ROD, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics 28:2537–2539
Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Razafinarivo NJ, Guyot R, Davis AP et al (2013) Genetic structure and diversity of coffee (Coffea) across Africa and the Indian Ocean islands revealed using microsatellites. Ann Bot 111:229–248
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tang JF, Vosman B, Voorrips RE et al (2006) QualitySNP: a pipeline for detecting single nucleotide polymorphisms and insertions/deletions in EST data from diploid and polyploid species. BMC Bioinform 7:438
Tshilenge P, Nkongolo KK, Mehes M et al (2009) Genetic variation in Coffea canephora L. (var. Robusta) accessions from the founder gene pool evaluated with ISSR and RAPD. Afr J Biotechnol 8:380–390
Tsuda T, Yamaguchi M, Honda C, Moriguchi T (2004) Expression of anthocyanin biosynthesis genes in the epicarp of peach and nectarine fruit. J Am Soc Hortic Sci 129:857–862
Várzea VMP, Marques VD, Pereira AP, Silva MC (2009) The use of Sarchimor derivatives in coffee breeding resistance to leaf rust. 22nd International Conference on Coffee Science, ASIC 2008, Campinas, SP, Brazil, 14–19 September, 2008 pp. 1424–1429
Vega FE (2008) The rise of coffee. Am Sci 96:138–145
Vega FE, Ebert A, Ming R (2008) Coffee germplasm resources, genomics, and breeding. Plant Breed Rev 30:415–447
Vidal RO, Mondego JMC, Pot D et al (2010) A high-throughput data mining of single nucleotide polymorphisms in Coffea species expressed sequence tags suggests differential homeologous gene expression in the allotetraploid Coffea arabica. Plant Physiol 154:1053–1066
Vieira LGE, Carvalho AA, Augusto CC et al (2006) Brazilian coffee genome project: an EST-based genomic resource. Braz J Plant Physiol 18:95–108
Vieira ESN, Von Pinho EVR, Carvalho MGG et al (2010) Development of microsatellite markers for identifying Brazilian Coffea arabica varieties. Genet Mol Biol 33:507–514
Wang B, Tan H, Fang W et al (2015) Developing single nucleotide polymorphism (SNP) markers from transcriptome sequences for identification of longan (Dimocarpus longan) germplasm. Hortic Res 2:14065
Wu GA, Prochnik S, Jenkins J et al (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 32:656–662
Yuyama PM, Júnior OR, Ivamoto ST et al (2016) Transcriptome analysis in Coffea eugenioides, an Arabica coffee ancestor, reveals differentially expressed genes in leaves and fruits. Mol Genet Genomics 291:323–336
Zhang D, Mischke S, Goenaga R et al (2006) Accuracy and reliability of high-throughput microsatellite genotyping for cacao individual identification. Crop Sci 46:2084–2092
Acknowledgments
We would like to give special thanks to Stephen Pinney (USDA, ARS) for assistance with SNP genotyping using the nanofluidic array system. References by USDA to a company and/or product are only for the purposes of information and do not imply approval or recommendation of the product to the exclusion of others that may also be suitable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Communicated by: Alan Andrade
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
(XLS 242 kb)
Supplemental Table 2
(XLSX 28 kb)
Rights and permissions
About this article
Cite this article
Zhou, L., Vega, F.E., Tan, H. et al. Developing Single Nucleotide Polymorphism (SNP) Markers for the Identification of Coffee Germplasm. Tropical Plant Biol. 9, 82–95 (2016). https://doi.org/10.1007/s12042-016-9167-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-016-9167-2